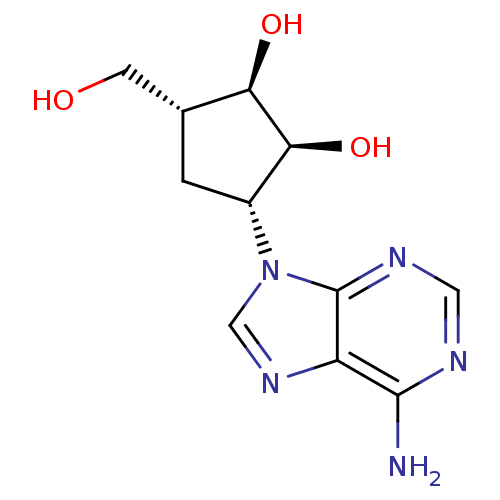
BDBM50088426 (1R,2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol::3-(6-Amino-purin-9-yl)-5-hydroxymethyl-cyclopentane-1,2-diol::CHEMBL49935
SMILES Nc1ncnc2n(cnc12)[C@@H]1C[C@H](CO)[C@@H](O)[C@H]1O
InChI Key InChIKey=UGRNVLGKAGREKS-GCXDCGAKSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50088426
Found 2 hits for monomerid = 50088426
Affinity DataKi: 2.00E+4nMAssay Description:Displacement of [125I]- AB-MECA from rat adenosine A3 receptor expressed in CHO cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.00E+5nMAssay Description:Binding affinity determined by displacement of specific binding of [125I]N-(4-amino-3-iodophenethyl)-adenosine in membranes of CHO cells stably trans...More data for this Ligand-Target Pair